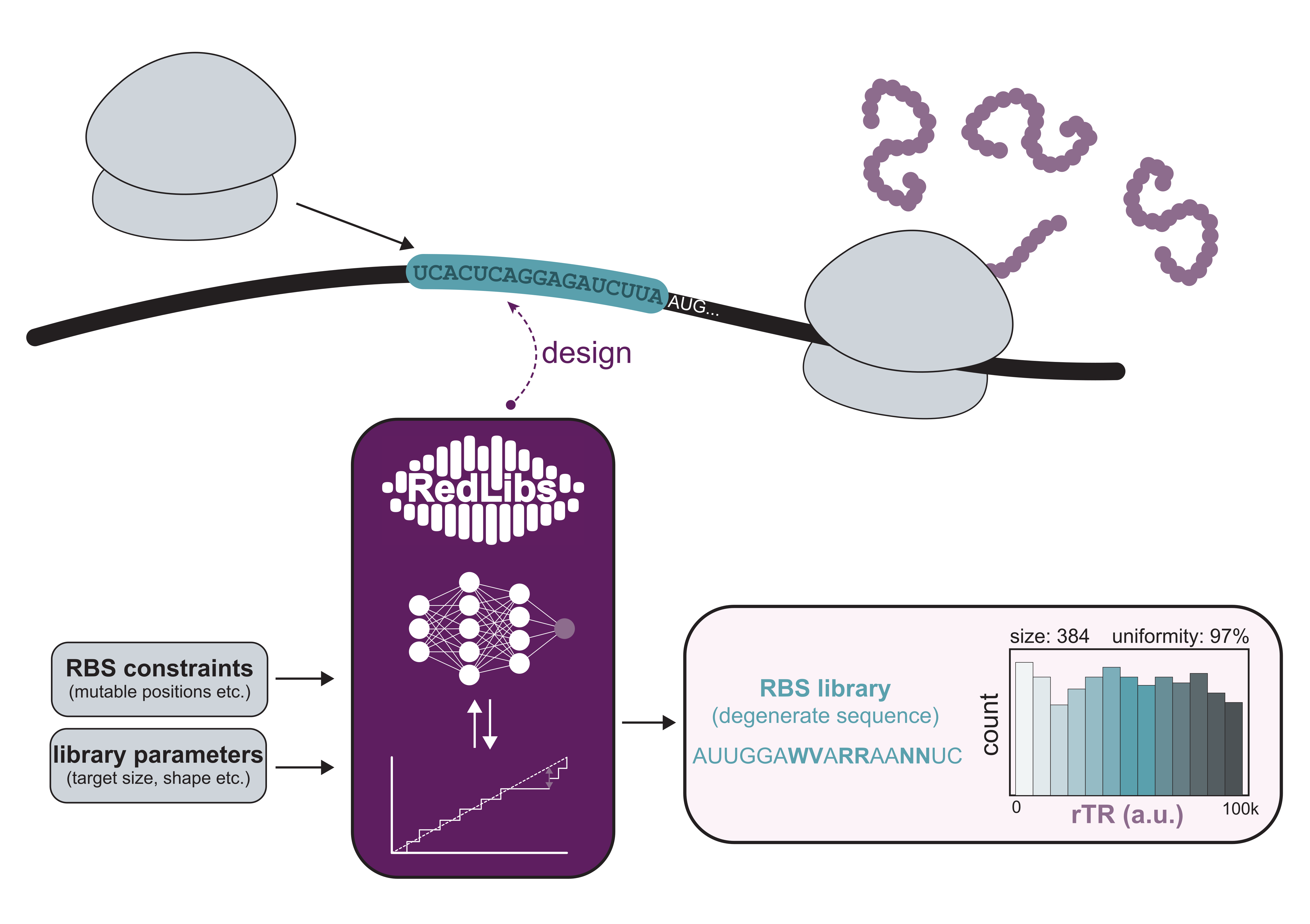
RedLibs is a tool that generates
RBS libraries
encoded by individual degenerate sequences
for straight-forward cloning.
These libraries have a user-defined size (i.e. number of RBS variants encoded by the degenerate sequence), and span the
rTR
range from low to high following a uniform distribution to optimally “scan” a range of different expression levels.
User manual
RedLibs uses prediction data derived from RBS Predictor to design RBS libraries following different sequence and library constraints
defined by the user.
First input mask (sequence constraints)
First, different sequence constraints are specified that define the mutational freedom and desired restrictions under which the subsequent library
optimization is taking place. Under Template, the user must specify the
5’-UTR
sequence from which the optimization should start (17 nts upstream of the start codon). In most cases, this should be the sequence in the parent
genetic construct (e.g. plasmid) that is to be optimized with the designed library. Via Mutable positions, the user then indicates which
positions in the template may be mutated as part of the library optimization. Positions are earmarked as mutable with N (full mutational freedom)
or any other degenerate base following the
IUPAC code.
Example:
| Template: |
AATATCTTAGCTAAATA[ATG] |
| Mutable positions: |
AATATNRRNNNTAAATA |
→
Result: RedLibs will suggest libraries allowing mutations only in the underlined positions of the template. N: A, C, T or G allowed.
R: A or G allowed. The [ATG] start codon must not be added to the query.
While adjusting Template and Mutable positions will suffice for most purposes, RedLibs offers the possibility to add further
sequence constraints via the dropdown Advanced options. Here, the user can further specify the maximum number of mutations and the maximum
number of consecutive mutations that should appear in the library relative to the template (see mouseovers for further information).
Second input mask (library constraints)
Next, different target parameters for the designed RBS libraries can be specified. The only required parameter here is the Target library size,
which corresponds to the number of individual RBS sequences (i.e. library members) that the designed library will contain. This parameter defines the
resolution (i.e. the number of discrete levels), with which the
rTR
range from 0 to 100,000 a.u. will be covered. It should be selected in agreement with the experimental throughput, at which library members can be
tested afterwards.
Example:
| Template: |
AATATCTTAGCTAAATA[ATG] |
| Mutable positions: |
AATATNRRNNNTAAATA |
| Target library size: |
18 |
→
Result: RedLibs will suggest libraries with a size of 18 RBS variants encoded by single, degenerate sequences following the sequence
constraints defined in the previous window.
Under Advanced options, additional library parameters can be defined (see mouseovers for detailed information). Lower/Upper rTR threshold
may be used to set lower and upper boundaries for the rTR range that should be uniformly covered (default: 0 – 100,000 a.u.). No. of top solutions
defines the number of top libraries that RedLibs suggests in the output (default: 10).
Output
The output of RedLibs consists of two files detailing the results of library optimization in a text-based (*.txt) and graphical (*.png) form.
The main output file (*.txt) is a comma-separated list of the optimized RBS libraries in order of decreasing
uniformity
It contains key sequence and numerical parameters of the top libraries and their individual members. Each individual library member is represented by
one line with four columns specifying its sequence (“# sequence”) and predicted rTR (“rTR_(a.u.)”) as well as the degenerate sequence
(“library”) and size (“size”) of the library it belongs to. A fifth column provides the
uniformity
(“uniformity_(%)”) as a score indicative of library quality with 100% corresponding to a perfectly uniform library matching the target distribution.
The graphical output (*.png) additionally visualizes key features of the top libraries. It entails a histogram representation of the rTR distribution
including a header containing the degenerate sequence, size and uniformity score for each library.
Example (*.txt):
| # sequence,rTR_(a.u.),library,size,uniformity_(%) |
|
| AATATTGGTGTTAAATA,3265.529,AATATWGGDGDTAAATA,18,90.9 |
[1] |
| AATATAGGTGTTAAATA,22384.644,AATATWGGDGDTAAATA,18,90.9 |
[2] |
| AATATTGGGGTTAAATA,32109.827, AATATWGGDGDTAAATA,18,90.9 |
[3] |
| ... |
|
| AATATAGGAGATAAATA,71568.020,AATATWGGDGDTAAATA,18,90.9 |
[18] |
| AATATTGGGTGTAAATA,8708.594,AATATDGGRDGTAAATA,18,89.9 |
[19] |
| AATATGGGGTGTAAATA,44077.774,AATATDGGRDGTAAATA,18,89.9 |
[20] |
| AATATAGGGTGTAAATA,29760.909,AATATDGGRDGTAAATA,18,89.9 |
[21] |
| ... |
|
| AATATAGGAAGTAAATA,24727.108,AATATDGGRDGTAAATA,18,89.9 |
[36] |
| ... |
|
→
In this example, a target library size of 18 was applied. Thus, the first 18 entries are members of the Top1 library, which share
the same degenerate sequence and uniformity score. Top2 library: entries 19-36, Top3 library: entries 37-54, etc..
Example (*.png):
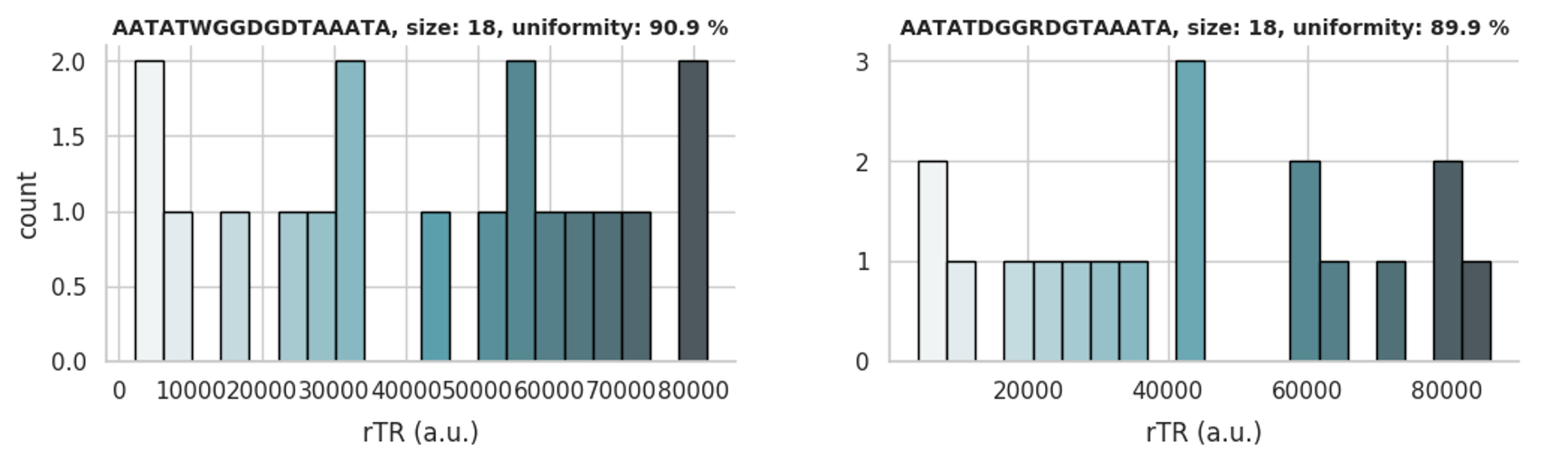
→
In this example, the graphical representation of two top libraries with a size of 18 is shown. The libraries are encoded by
sequences AATATWGGDGDTAAATA and AATATDGGRDGTAAATA, respectively, and cover the rTR range
between 0 and 100,000 a.u. with good uniformity.
When you use this pipeline in your published work, please do not forget to cite:
- Jeschek, M., Gerngross, D., & Panke, S. (2016).
Rationally reduced libraries for combinatorial pathway optimization minimizing experimental effort.
Nature communications, 7, 11163
(https://doi.org/10.1038/ncomms11163)
- Höllerer, S., Papaxanthos, L., Gumpinger, A. C., Fischer, K., Beisel, C., Borgwardt, K., Benenson, Y., & Jeschek, M. (2020).
Large-scale DNA-based phenotypic recording and deep learning enable highly accurate sequence-function mapping.
Nature communications, 11(1), 3551
(https://doi.org/10.1038/s41467-020-17222-4)